Code usage
usage.Rmd1 Convention
Purpose of groups of functions in the package:
- calculate_* - calculates one specific value. e.q. deuterium uptake values for specific time point.
- create_* - creates a dataset for set of values e.q. multiple time points.
- plot_* - plots a visualization of provided data, accepts calculate_* and create_* outputs.
- show_* - subsets the dataset to show specific values, accepts calculate_* and create_* outputs.
Variables:
- deut_uptake - deuterium uptake [Da]
- frac_deut_uptake - fractional deuterium uptake [%]
- theo_deut_uptake - theoretical deuterium uptake [Da]
- theo_frac_deut_uptake - theoretical fractional deuterium uptake [%]
- diff_deut_uptake - differential deuterium uptake [Da]
- frac_diff_deut_uptake - theoretical differential fractional deuterium uptake [%]
- theo_diff_deut_uptake - differential deuterium uptake [Da]
- theo_frac_diff_deut_uptake - theoretical differential fractional deuterium uptake [%]
2 Example of use
2.1 Read data file
dat <- read_hdx(system.file(package = "HaDeX2", "HaDeX/data/alpha.csv"))2.2 Different forms of visualization
Create data frame with uptake values for state in all time points:
uptake_dat <- create_state_uptake_dataset(dat)Butterfly:
plot_butterfly(uptake_dat)
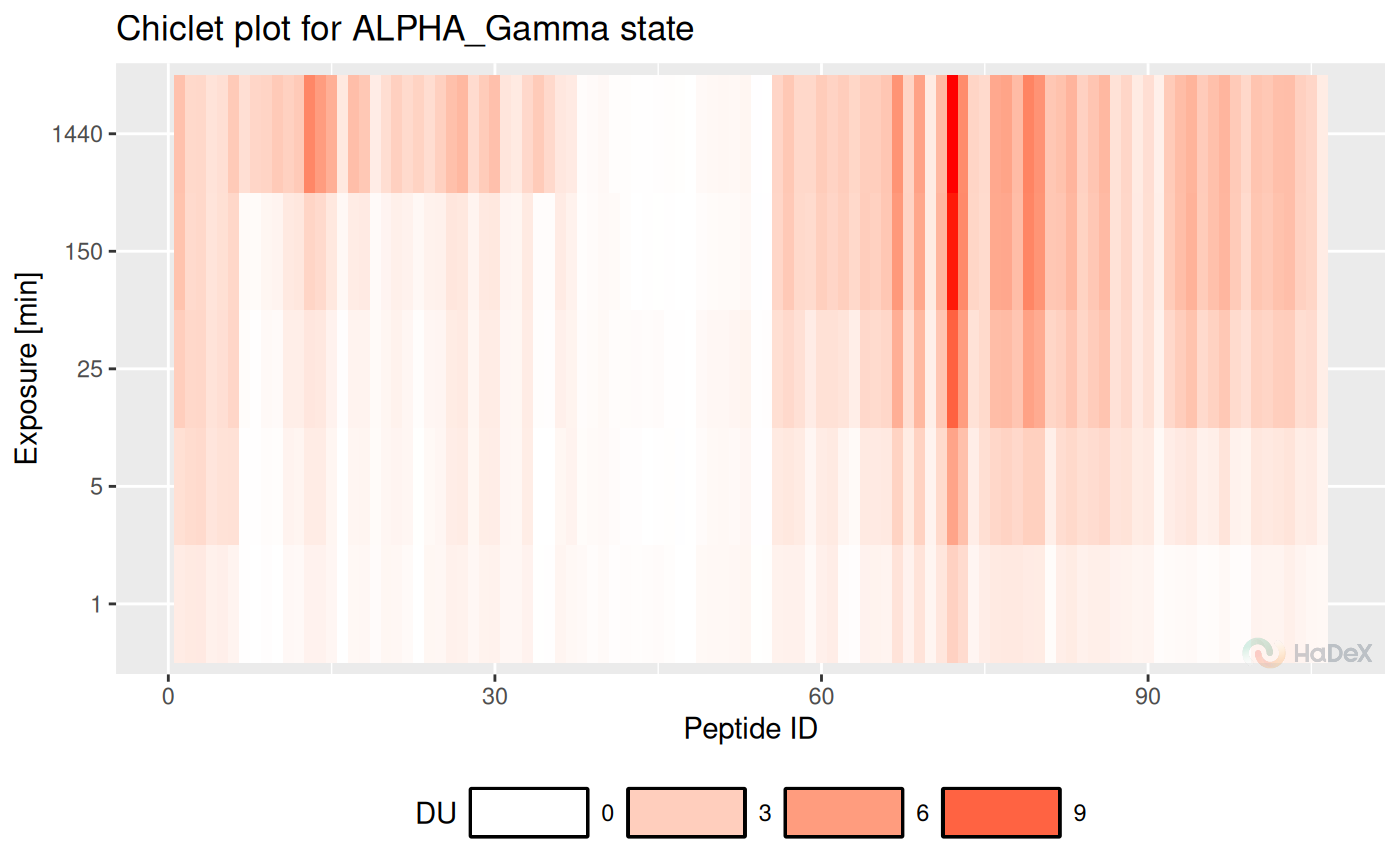
Chiclet:
plot_chiclet(uptake_dat)## Warning: The `size` argument of `element_rect()` is deprecated as of ggplot2 3.4.0.
## ℹ Please use the `linewidth` argument instead.
## ℹ The deprecated feature was likely used in the HaDeX2 package.
## Please report the issue to the authors.
## This warning is displayed once per session.
## Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
## generated.