Coverage heatmap
plot_coverage_heatmap.RdCoverage heatmap with color indicating specific value
Usage
plot_coverage_heatmap(
x_dat,
protein = x_dat[["Protein"]][1],
state = NULL,
value = NULL,
time_t = NULL,
interactive = getOption("hadex_use_interactive_plots")
)Arguments
- x_dat
data created using calculate_ or create_ function
- protein
selected protein
- state
selected biological state
- value
value to be presented
- time_t
chosen time point
- interactive
logical, whether plot should have an interactive layer created with with ggiraph, which would add tooltips to the plot in an interactive display (HTML/Markdown documents or shiny app)
Value
a ggplot object
Examples
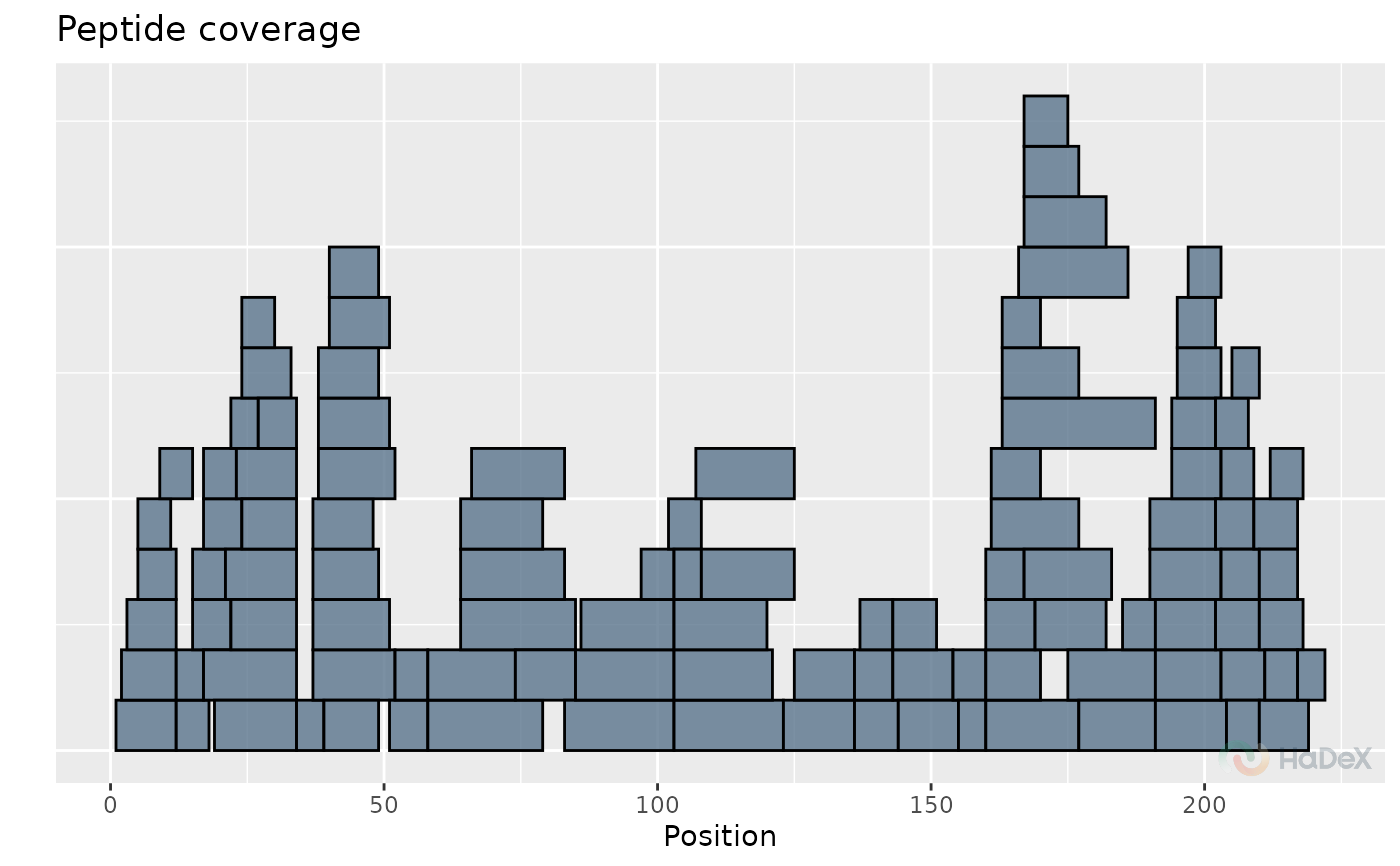
uptake_dat <- create_uptake_dataset(alpha_dat, states = "Alpha_KSCN")
plot_coverage_heatmap(uptake_dat)
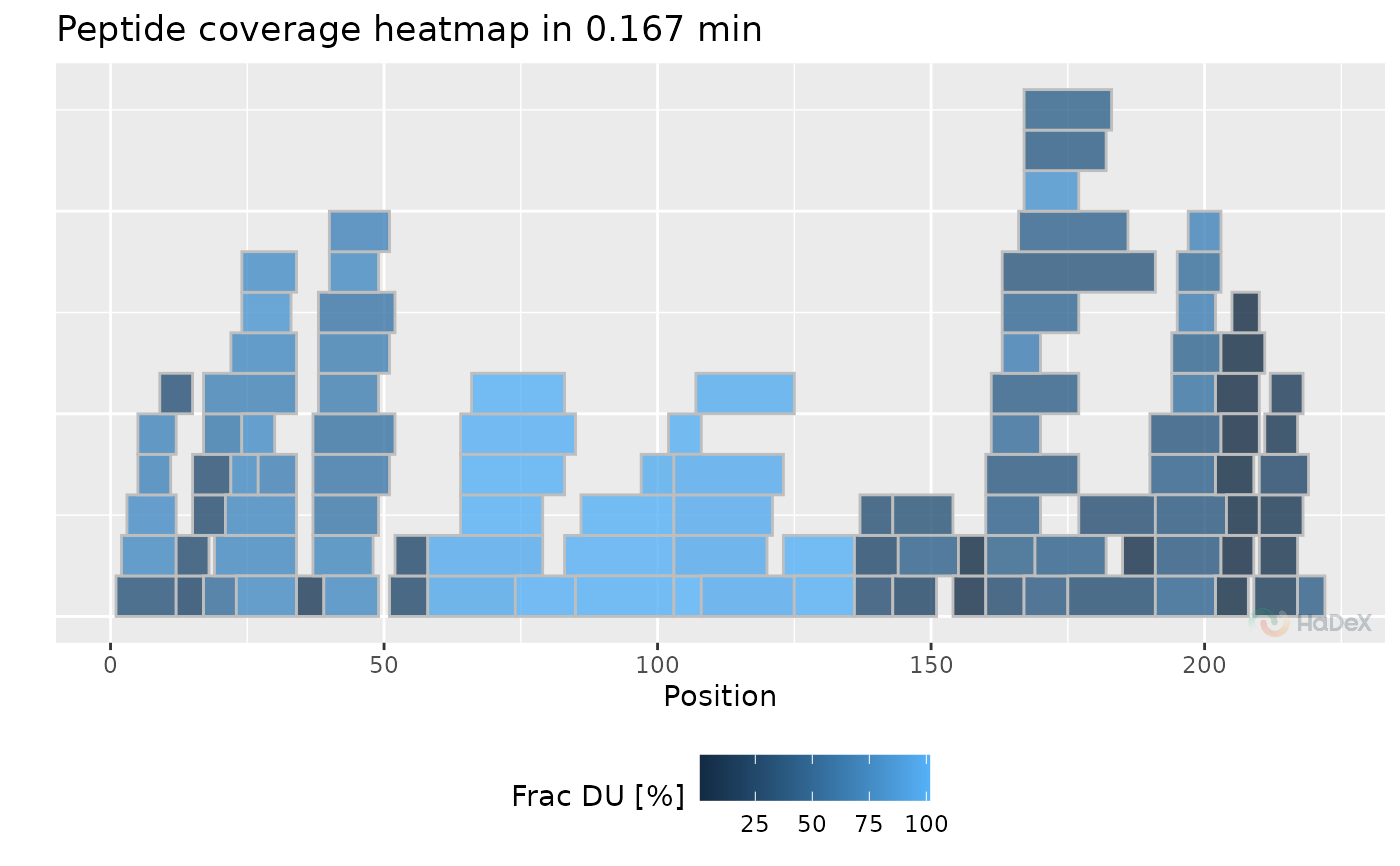
 plot_coverage_heatmap(x_dat = uptake_dat, value = "frac_deut_uptake", time_t = 0.167)
#> Warning: `aes_string()` was deprecated in ggplot2 3.0.0.
#> ℹ Please use tidy evaluation idioms with `aes()`.
#> ℹ See also `vignette("ggplot2-in-packages")` for more information.
#> ℹ The deprecated feature was likely used in the HaDeX2 package.
#> Please report the issue to the authors.
#> Ignoring unknown labels:
#> • colour : "Exposure"
plot_coverage_heatmap(x_dat = uptake_dat, value = "frac_deut_uptake", time_t = 0.167)
#> Warning: `aes_string()` was deprecated in ggplot2 3.0.0.
#> ℹ Please use tidy evaluation idioms with `aes()`.
#> ℹ See also `vignette("ggplot2-in-packages")` for more information.
#> ℹ The deprecated feature was likely used in the HaDeX2 package.
#> Please report the issue to the authors.
#> Ignoring unknown labels:
#> • colour : "Exposure"
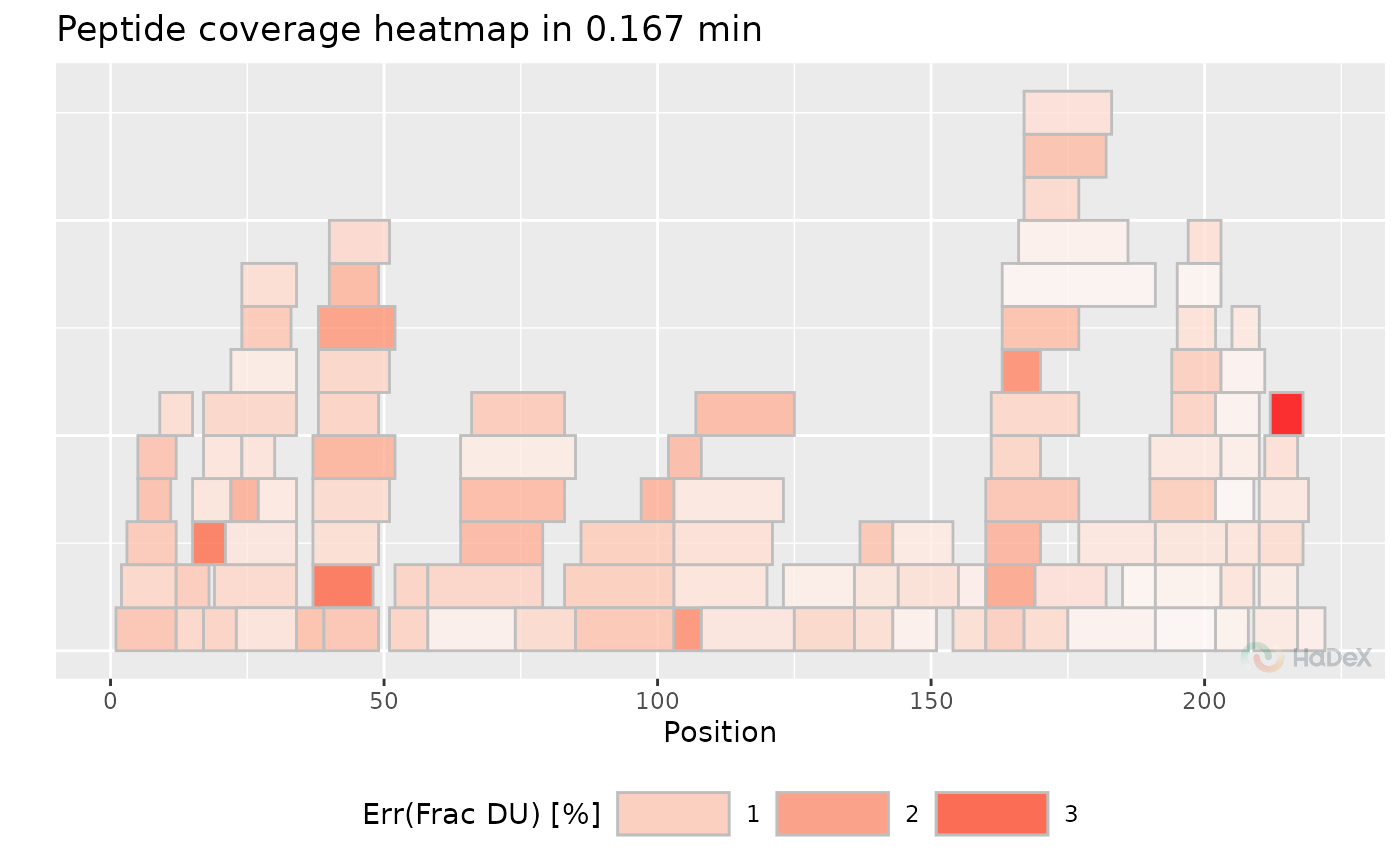
 plot_coverage_heatmap(uptake_dat, value = "err_frac_deut_uptake", time_t = 0.167)
#> Ignoring unknown labels:
#> • colour : "Exposure"
plot_coverage_heatmap(uptake_dat, value = "err_frac_deut_uptake", time_t = 0.167)
#> Ignoring unknown labels:
#> • colour : "Exposure"
 diff_uptake_dat <- create_diff_uptake_dataset(alpha_dat)
plot_coverage_heatmap(diff_uptake_dat)
diff_uptake_dat <- create_diff_uptake_dataset(alpha_dat)
plot_coverage_heatmap(diff_uptake_dat)
 plot_coverage_heatmap(diff_uptake_dat, value = "diff_frac_deut_uptake")
#> No time point selected!
plot_coverage_heatmap(diff_uptake_dat, value = "diff_frac_deut_uptake")
#> No time point selected!
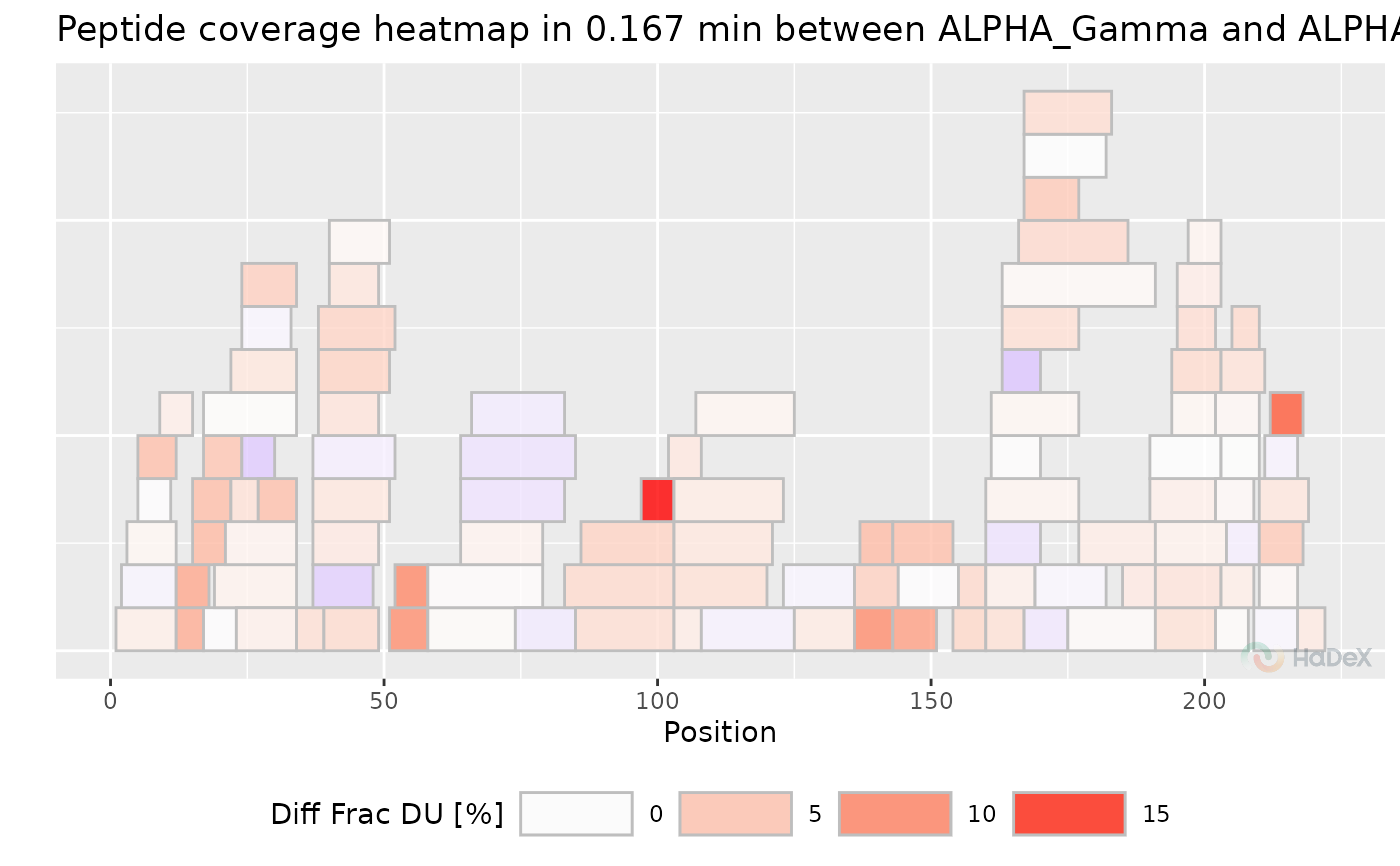
 plot_coverage_heatmap(diff_uptake_dat, value = "diff_frac_deut_uptake", time_t = 0.167)
#> Ignoring unknown labels:
#> • colour : "Exposure"
plot_coverage_heatmap(diff_uptake_dat, value = "diff_frac_deut_uptake", time_t = 0.167)
#> Ignoring unknown labels:
#> • colour : "Exposure"
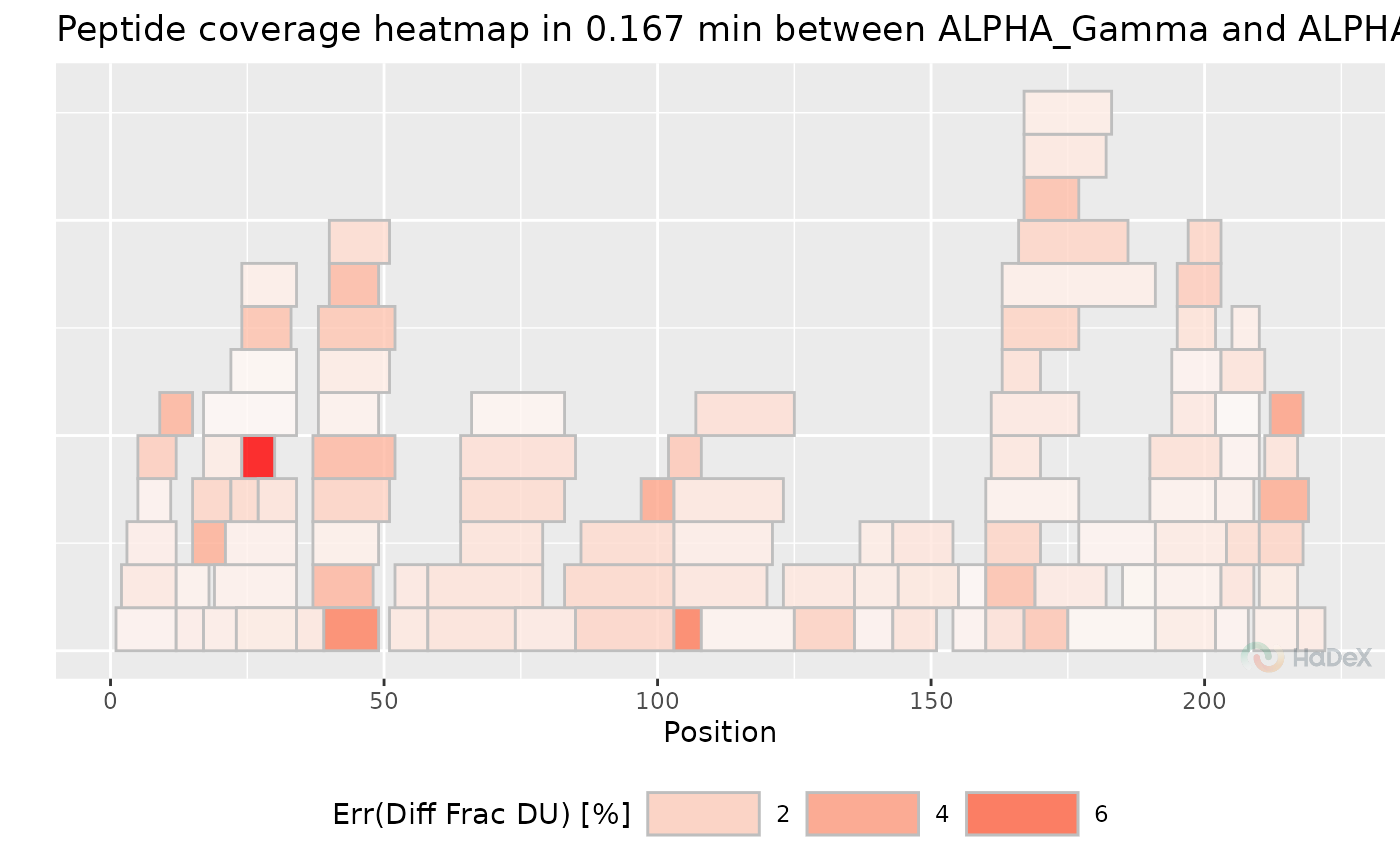
 plot_coverage_heatmap(diff_uptake_dat, value = "err_diff_frac_deut_uptake", time_t = 0.167)
#> Scale for fill is already present.
#> Adding another scale for fill, which will replace the existing scale.
#> Ignoring unknown labels:
#> • colour : "Exposure"
plot_coverage_heatmap(diff_uptake_dat, value = "err_diff_frac_deut_uptake", time_t = 0.167)
#> Scale for fill is already present.
#> Adding another scale for fill, which will replace the existing scale.
#> Ignoring unknown labels:
#> • colour : "Exposure"
 auc_dat <- calculate_auc(create_uptake_dataset(alpha_dat))
plot_coverage_heatmap(auc_dat, value = "auc")
#> Ignoring unknown labels:
#> • colour : "Exposure"
auc_dat <- calculate_auc(create_uptake_dataset(alpha_dat))
plot_coverage_heatmap(auc_dat, value = "auc")
#> Ignoring unknown labels:
#> • colour : "Exposure"
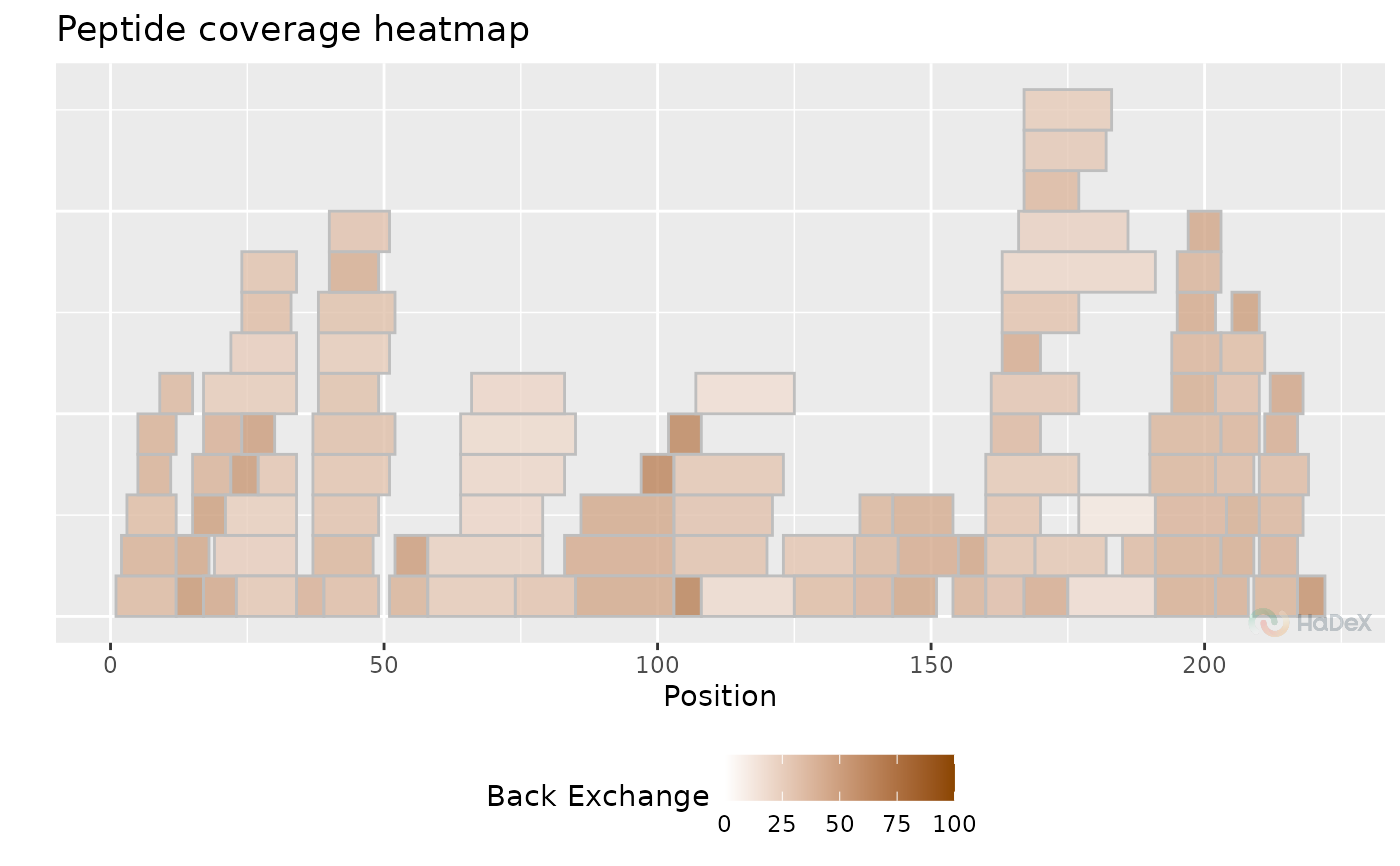
 bex_dat <- calculate_back_exchange(alpha_dat, state = "Alpha_KSCN")
plot_coverage_heatmap(bex_dat, value = "back_exchange")
#> Ignoring unknown labels:
#> • colour : "Exposure"
bex_dat <- calculate_back_exchange(alpha_dat, state = "Alpha_KSCN")
plot_coverage_heatmap(bex_dat, value = "back_exchange")
#> Ignoring unknown labels:
#> • colour : "Exposure"