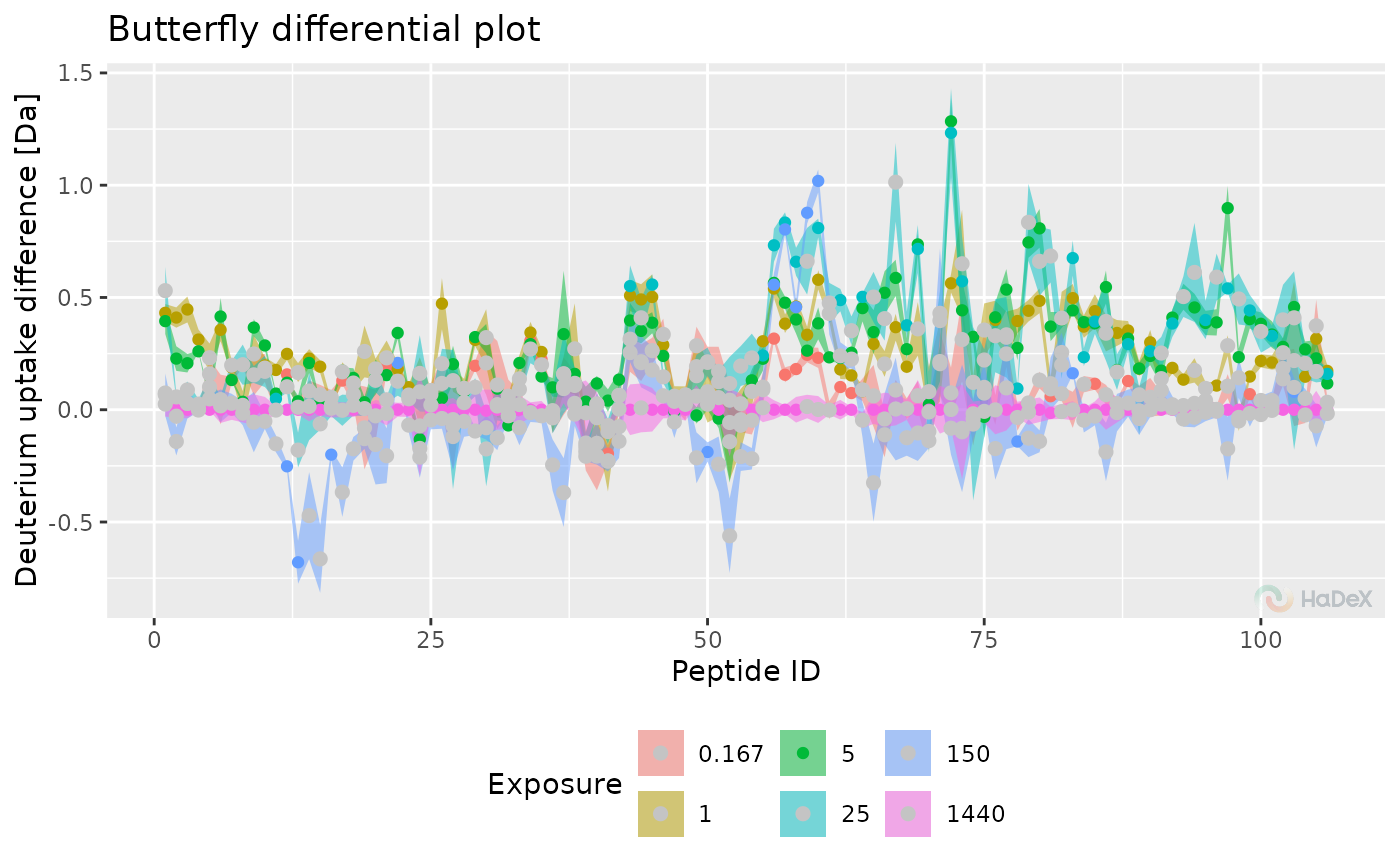
Butterfly differential deuterium uptake plot
plot_differential_butterfly.RdButterfly plot of differential deuterium uptake values between two biological states in time.
Usage
plot_differential_butterfly(
diff_uptake_dat = NULL,
diff_p_uptake_dat = NULL,
theoretical = FALSE,
fractional = FALSE,
show_houde_interval = FALSE,
show_tstud_confidence = FALSE,
uncertainty_type = "ribbon",
confidence_level = 0.98,
interactive = getOption("hadex_use_interactive_plots")
)Arguments
- diff_uptake_dat
data produced by
calculate_diff_uptakeorcreate_diff_uptake_datasetfunction- diff_p_uptake_dat
differential uptake data alongside the P-value as calculated by
create_p_diff_uptake_dataset- theoretical
logical, determines if values are theoretical- fractional
logical, determines if values are fractional- show_houde_interval
logical, determines if houde interval is shown- show_tstud_confidence
logical, determines if t-Student test validity is shown- uncertainty_type
type of presenting uncertainty, possible values: "ribbon", "bars" or "bars + line"
- confidence_level
confidence level for the test, from range [0, 1] Important if selected show_confidence_limit
- interactive
logical, whether plot should have an interactive layer created with with ggiraph, which would add tooltips to the plot in an interactive display (HTML/Markdown documents or shiny app).
Details
Function plot_differential_butterfly generates
differential butterfly plot based on provided data and parameters. On X-axis
there is peptide ID. On the Y-axis there is deuterium uptake difference in
chosen form. Data from multiple time points of measurement is presented.
If chosen, there are confidence limits based on Houde test on provided
confidence level.
References
Houde, D., Berkowitz, S.A., and Engen, J.R. (2011). The Utility of Hydrogen/Deuterium Exchange Mass Spectrometry in Biopharmaceutical Comparability Studies. J Pharm Sci 100, 2071–2086.
Examples
diff_uptake_dat <- create_diff_uptake_dataset(alpha_dat)
plot_differential_butterfly(diff_uptake_dat = diff_uptake_dat)
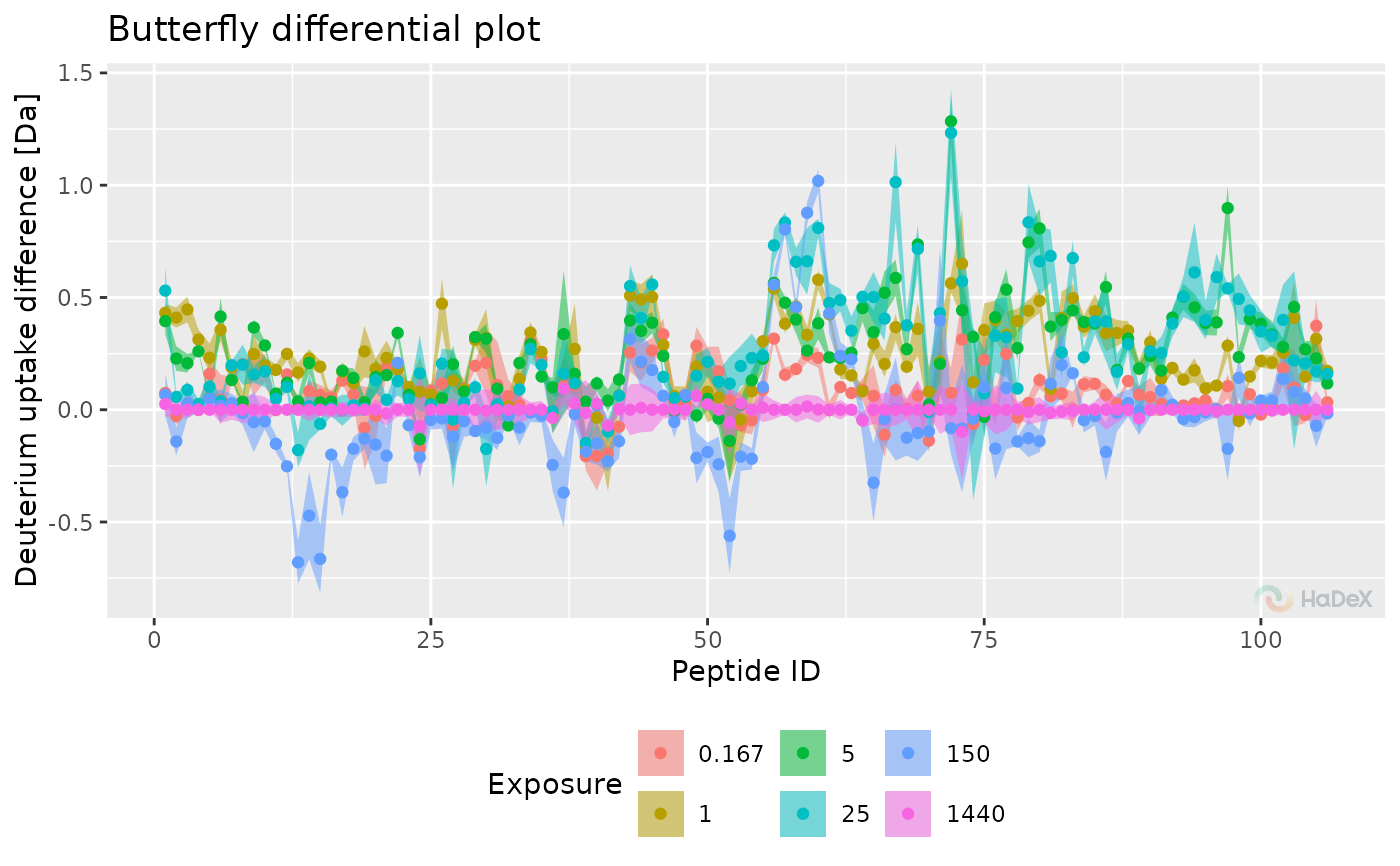 plot_differential_butterfly(diff_uptake_dat = diff_uptake_dat, show_houde_interval = TRUE)
plot_differential_butterfly(diff_uptake_dat = diff_uptake_dat, show_houde_interval = TRUE)
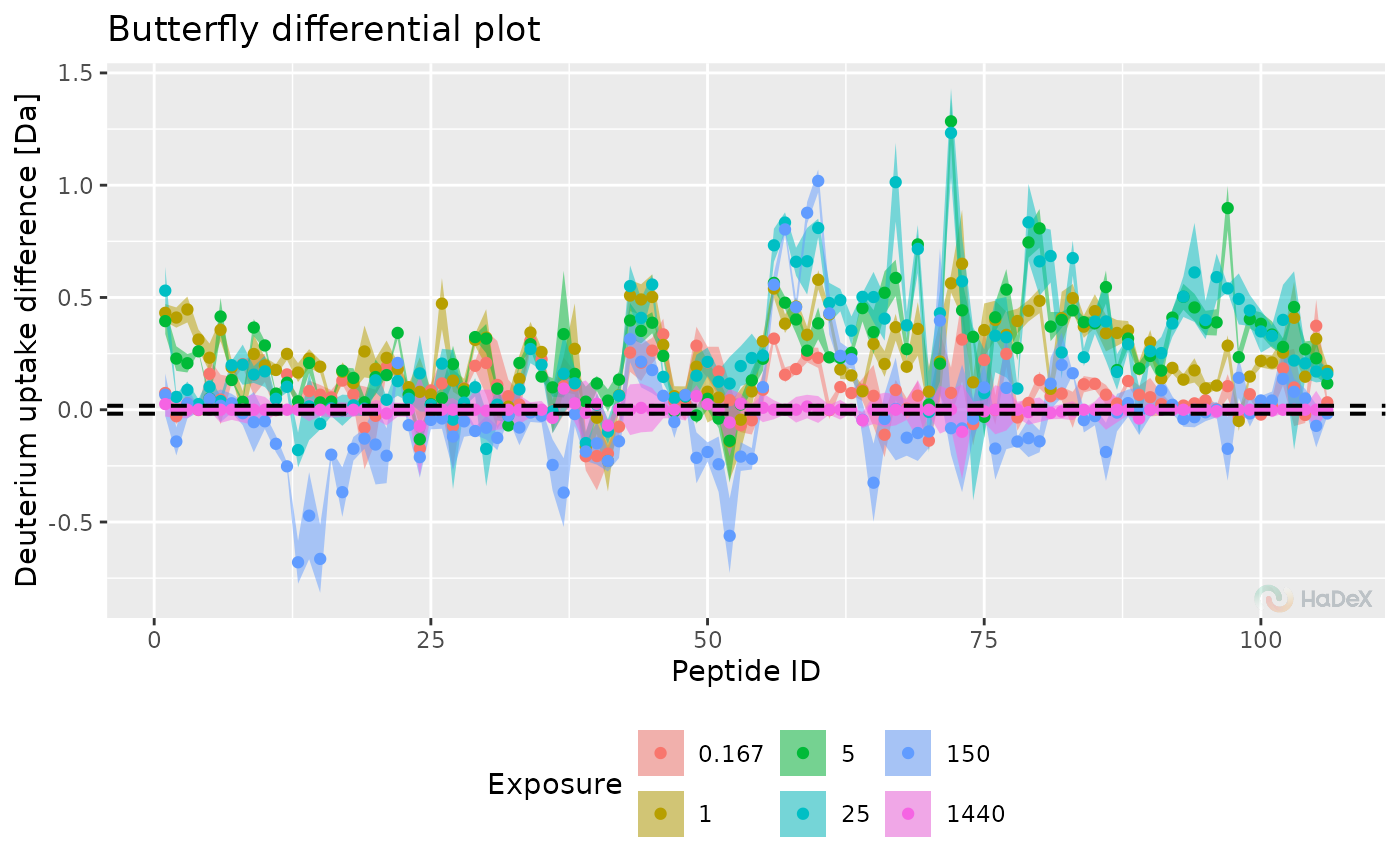 diff_p_uptake_dat <- create_p_diff_uptake_dataset(alpha_dat)
plot_differential_butterfly(diff_p_uptake_dat = diff_p_uptake_dat, show_tstud_confidence = TRUE)
diff_p_uptake_dat <- create_p_diff_uptake_dataset(alpha_dat)
plot_differential_butterfly(diff_p_uptake_dat = diff_p_uptake_dat, show_tstud_confidence = TRUE)